HighScriber™ - a blend of Reverse Transcriptase and RNase Inhibitor for an efficient cDNA synthesis
HighScriber™ - a blend of Reverse Transcriptase and RNase Inhibitor for an efficient cDNA synthesis
Applications
- cDNA synthesis of up to 15 kb long transcripts.
- Template generation for RT-PCR & RT-qPCR.
- cDNA synthesis from complex templates.
Benefits
- Thermostable Reverse Transcriptase blended with Ribonuclease Inhibitor for efficient cDNA synthesis.
- High yields of full lengths transcripts up to 12-15 kb.
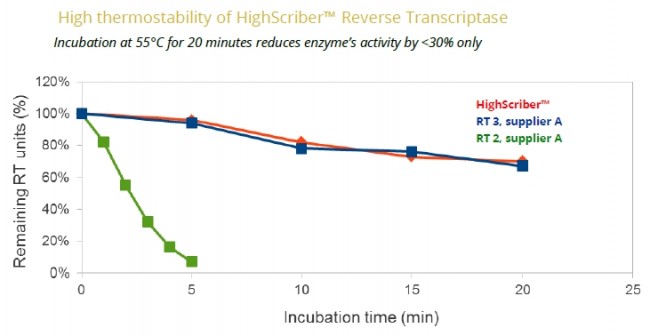
- cDNA synthesis from complex templates at up to 55°C.
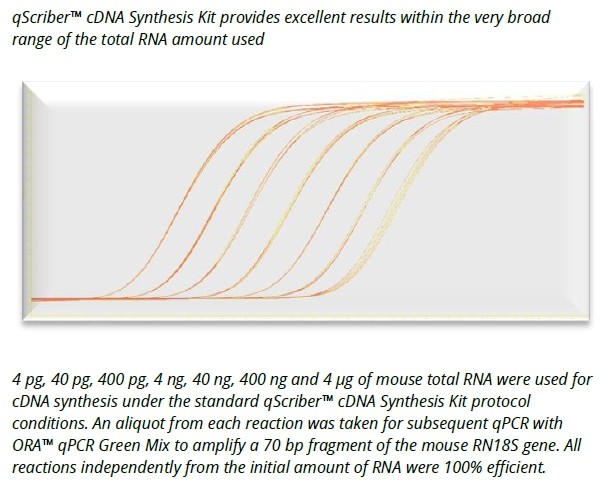
- High sensitivity detection from 1 pg of total RNA template.
HighScriber™ - a blend of Reverse Transcriptase and RNase Inhibitor for an efficient cDNA synthesis
HighScriber™ - a blend of Reverse Transcriptase and RNase Inhibitor for an efficient cDNA synthesis










- Description The HighScriber™ Reverse Transcriptase Mix is a premium tool for the high efficiency reverse transcription of up to 12-15 kb… More
- Protocols Download Protocol and Specifications - Product Insert HighScriber™ Reverse Transcriptase Mix, 20X Ask for a Samp More
- Specifications Download Protocol and Specifications - Product Insert HighScriber™ Reverse Transcriptase Mix, 20X Download MSDS& More
- Resources Download Protocol and Specifications - Product Insert HighScriber™ Reverse Transcriptase Mix, 20X Download MSDS& More
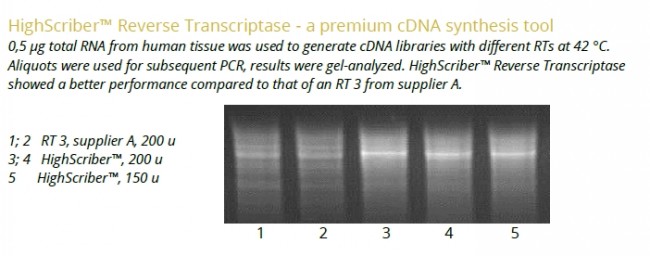
The HighScriber™ Reverse Transcriptase Mix is a premium tool for the high efficiency reverse transcription of up to 12-15 kb long cDNA. Mix includes HighScriber™ Reverse Transcriptase and Ribonuclease Inhibitor for save, robust cDNA synthesis and ease of use. HighScriber™ Reverse Transcriptase allows for high detection sensitivity from 1 pg of total RNA. The wide reaction temperature range (38°C - 55°C) ensures efficient cDNA synthesis from complex or GC rich templates.
Choosing the right reverse transcriptase
Different reverse transcriptase used for cDNA synthesis have different properties and therefore not all of them are suitable for the same applications. Here are some important factors to consider when choosing the right reverse transcriptase:
• Some reverse transcriptases work at lower temperatures, such as 42°C (MMuLV RT), while thermostable reverse transcriptases work at higher temperatures, such as 50°C or 55°C (highQu HighScriber™ RT). The more complex templates are transcribed, the higher temperature is recommended to disrupt secondary structures of the RNA.
• Reverse transcriptases might have template preferences and are more efficient at synthesizing cDNA from certain RNA templates, such as mRNA, while others can synthesize cDNA from a wider range of RNA templates.
• The sensitivity of a reverse transcriptase becomes especially important if the RNA sample is of low copy-number concentration. Not all enzymes can synthesize cDNA from low amounts of RNA efficiently.
• The speed of reverse transcriptases also differs. Some RT enzymes work faster than others and ensure high cDNA synthesis yield in less time.
Good enzymes are typically supplied with optimized reverse transcription reagents to ensure high reverse transcription yields, enable high temperature protocols, provide high specificity and unbiased cDNA synthesis.
- Download Protocol and Specifications - Product Insert HighScriber™ Reverse Transcriptase Mix, 20X
- Ask for a Sample today
- Have technical questions? Contact us
- Download a list of Publications mentioning highQu products
- Or see how others use our products at bioz.com or google scholar
- Download Protocol and Specifications - Product Insert HighScriber™ Reverse Transcriptase Mix, 20X
- Download MSDS HighScriber™ Reverse Transcriptase Mix, 20X
- Need a lot-specific Certificate of Analysis? E-mail us at info@highQu.com
- Want custom formulations or bulk sizes? E-mail us at info@highQu.com, and check our OEM offers
- Have more specific questions? Contact us
- Download Protocol and Specifications - Product Insert HighScriber™ Reverse Transcriptase Mix, 20X
- Download MSDS HighScriber™ Reverse Transcriptase Mix, 20X
- Download highQu Catalogue of Premium Research Tools
- Download a list of Publications mentioning highQu products
- Or see how others use our products at bioz.com or google scholar
- Download Product Pricelist (DE 2021)
- Download all highQu Product Inserts and MSDS sheets
- Have questions? Contact us
Related Products